Enhancing Precision Digital Pathology with an AI-powered Platform Accelerated by Supercomputers
Goals: We propose a new platform to jointly analyze digital pathology images demonstrated on rapidly evolving cancers from rare osteosarcoma (bone) cancer, squamous cell (skin) carcinoma, to adenocarcinoma (cervical, breast, colon, lung, liver, GI tract) – affecting not only the Vietnamese population but many U.S. and Vietnam veterans [1]. The novelty of our approach is that disease patterns learned from one patient cohort will successfully transfer and enhance the precision and understanding of another to comingle analytical applications on a shared AI-driven, supercomputer-accelerated platform [2, 3, 4] at Illinois. Leveraging the petascale processing power of NCSA’s world-class supercomputer [2] and CSL’s genomic accelerator [5], we will augment Vietnamese-anonymized patient pathology images and metadata1 with the comprehensive National Institutes of Health (NIH)’s 2.5 petabytes of The Cancer Genome Atlas (TCGA [6]) multi-omics data (20,000 primary cancer and matched normal samples spanning 33 cancer types), to create an accurate Vietnamese’s cancer atlas and the corresponding foundational AI prediction models, applicable on all high-demanding cancer types. Our technical contribution is to integrate complementary statistical methodologies, which involves: a) weakly supervised whole slice image segmentation; inference using label-free super-resolution images; c) early prediction of metastasis cancer by intelligently scheduling inference tasks using IBM’s POWER-9 microprocessor cluster at CSL-UIUC, while concurrently executing those tasks on a supercomputer of 40 flagship NVIDIA H100 GPUs [2] and an exclusive 400,000 AI-optimized cores wafer-scale appliance [7] at NCSA-UIUC. The platform will employ continuously evolving probabilistic graphical models pioneered by our PIs (e.g., Markov Random Fields [8] and Factor Graphs [9]) to alleviate uncertainty in the prediction of the above techniques while aiding the pathologist’s interpretability using Bayesian Networks [10, 11]. Using the above techniques, our platform will realize VinUni-Vinmec-Illinois-Mayo research breakthrough in the value-based healthcare domain, which has a substantial market demand (USD 3.2 trillion at a 7.5% annual growth rate).
Successful outcomes for key stakeholders are as follows: (1) This interdisciplinary project will create a platform to attract all-level students to VinUni, starting from high school, train undergrad/master/PhD students, and enrich supercomputing experience for postdoctoral scholars. VinUni researchers will gain exclusive computing allocation at the NCSA/CSL’s supercomputer platform while being guided by domain experts operating medical AI analytics at Mayo-NCSA’s secure HIPAA-compliant health enclave; (2) Vinmec will attain international renown through open-science Vietnamese cancer atlas and collect a sustained revenue stream of foundational AI pathology prediction models by licensing to U.S. hospitals focusing on Asian-American patients, earning royalties on a per-image-inference basis, or as a knowledge-base subscription service; (3) Vinmec’s AI-assisted pathologists will be productive and realize transformational clinical outcomes together with surgeons, e.g., early recognition of tumors in super-resolution whole-slice images. Most importantly, patient data will be safeguarded by HIPAA compliance technologies that are supported by our team’s cybersecurity AI secure health enclave (a separate proposal); (4) our collaborators from CSL-UIUC, NCSA-UIUC, Mayo will be able to extend their understanding of precision pathology research from a diverse, large-scale dataset of Vietnamese population. The proposed AI digital pathologist system (Fig. 1) will be deployed and operated by a team of anatomic/clinical pathologists at Vinmec, VISHC and CHS-VinUni immediately after successful completion of the first year.
Intellectual merits.
- Producing a repository of foundational AI models capable of reasoning on both rare (osteosarcoma) and widespread (gastrointestinal, cervical, squamous cells) cancers affecting the entire Vietnamese population and U.S. veterans who fought in the Vietnam wars. Co-construction of such AI models from scratch on the same platform allows shared multi-modal patient data, e.g., textual bi-lingual doctor notes, histology, and whole-slice images.
- Democratizing supercomputers as a centralized platform for collaborative inference of numerous AI-based computational pathology applications. Three representative computational pathology AI applications: osteosarcoma cancer, squamous cells on skin cancer, and gastrointestinal cancer screening using speech features [12], and polyp surveillance for colorectal cancer will be immediately evaluated on the platform in 2024 as we already processed data; with cervical cancer (25,000 diagnoses per year at Vinmec), prostate, colorectal, breast, stomach, and lung cancer will be gradually being rolled out (2025-2028).
- Inventing systems and methods for expedited pathology diagnosis by leveraging accelerated parallelism on IBM’s POWER micropro-cessor in collaboration with CSL-UIUC. The output research artifacts will be reproducible and reusable by other applications on a data lake that remains on-premise, jointly located at Illinois-Vinmec, and always accessible. Patient-sensitive data remains in Vietnam, while anonymized data donated for open-science research with the patient’s consent will be used for training at a supercomputer in NCSA/Illinois.
- Enabling open-science interdisciplinary evaluation of AI/ML/probabilistic diagnosis methods on urgent Vietnamese population problems. Our preliminary work through the CSL-NCSA-VinUni-Vinmec student exchange program has identified key bottlenecks in the care pathways of Osteosarcoma (bone) cancer that can be accelerated by AIs. Other Mayo collaborators, Vinmec Orthopedics & Sports Medicine motion lab, and VinUni faculty, such as the Advanced Imaging Education Center (AIEC), also confirmed their intent to work with us on training medical professionals in sports medicine imaging and colon cancer.
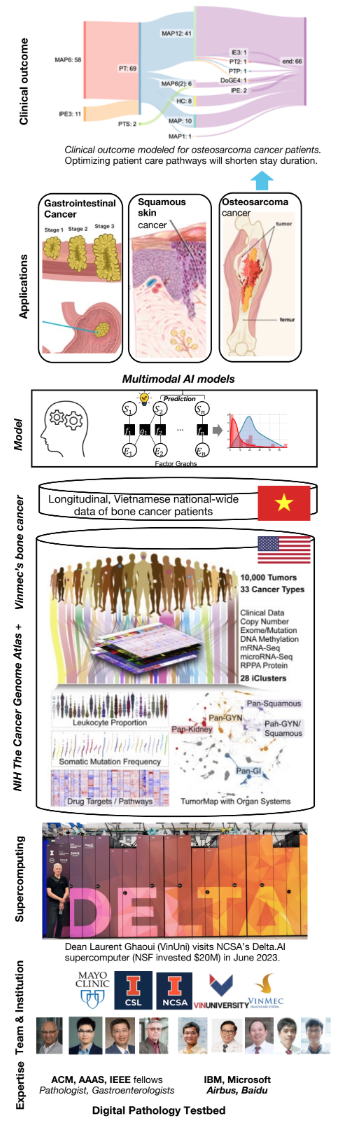
Figure 1: Supercomputing stack of the digital pathology platform and expected outcomes.
Broader impact. Our platform will support data analysis experiments of already awarded proposals and future applications. In addition, this proposal will drive a major economic im-pact by commercializing the above medical AI applications, platform, and subscription services while working closely with patients, policymakers, and educating the Vietnamese’s public to ensure environmental sustainability through green computing in collaboration with industry partners such as IBM. The important broader impact of our platform is to transform Vinmec’s quality of care and patient health outcomes, e.g., significantly lift the survival curve of the patients.
Student recruitment plan. The interdisciplinary nature of our strategic team (Mayo-Vinmec pathologists, VinUni surgeons, NCSA supercomputer operators, and imag-ing/machine learning/AI experts) has been and will continue to attract a large cohort of Vietnamese students with a diverse background from engineering to biomedicine. At UIUC, two VinUni students have successfully audited graduate-level AI/data science class by our lead PI (Prof. Iyer) and performed hands-on data analytics at NCSA through our NCSA – VISHC Exchange Research Program. Thereby, they are well-prepared for Illinois’ Ph.D. program and in-person collaboration with the Mayo Clinic through internships.
We have scheduled a special recruitment seminar in December with a panel of: 1) past VinUni and Illinois students discussing internship experiences, 2) Mayo doctors sharing experiences in the medical field, 3) Illinois faculty discussing research challenges. The seminar will connect with the SoICT conference in HCMC, with a broad attendance of students and healthcare experts. The seminar will be hybrid: led by VinUni’s PIs in person (Hanoi/HCMC) to interact with students and broadcast to interested VinUni students, and student selections will follow immediately. The future educational component of this proposal will be led by a closely-knit cohort of interdisciplinary experts (U.S.-educated with field experiences in Vinmec). We will train: five Ph.D.s (one with big data systems expertise, one with core statistical theories expertise, and three with different medical experiences, including digital pathologies, osteosarcoma cancer); three Masters (two medical systems engineering and one health informatics), and a minimum of ten under-graduate exchange students interning at Mayo Clinic, NCSA, CSL, and open-science U.S. national labs. Most importantly, interested microbiology pathologists at Vinmec can also participate in this unique PhD program to enhance their AI/computational skills.
Technology transfer plan. Our lead PI is leading the provisional patent process on key digital image pathology processing techniques in collaboration with Mayo. Any future techniques will be jointly patented between Mayo, VinUni, Vinmec, and Illinois upon mutual agreement by the end of the program. To accelerate technology transfer, we will consult the Technology Entrepreneur Center in Illinois with personalized advices from a leading Vietnamese-American financial technology expert with over 393 granted U.S. patents and Dr. Huyen Le, a U.S. Food and Drug Administration (FDA) fellow, to ensure our product’s compliance with federal regulations before going to the market.
Qualifications. Our team of leading academic experts (ACM, AAAS, and IEEE fellows) have rich industries (IBM, Microsoft, Airbus, and Baidu) and federal experiences (NSF Cybersecurity, NIH, NASA, NSA, DoE, and DoD). Much of our competency in statistics, image processing, High-Performance Computing security, AI, neural networks, and fed-erated learning have been developed through a nurturing relationships with Vietnamese students, particularly those from VinUni, in the past two decades. The next five years will mark our commitment to serving as dedicated mentors, leveraging our Illinois and VinUni’s expertise as the a force multiplier for their academic, professional, and Vinmec’s market growth.
External Fundings and long-term collaboration We expect this proposal will not only sustain itself but also support others to accelerate their experiments because of its in-terdisciplinary, predictive, personalized, precision, and participatory (4P) nature that will attract diverse funding sources. Existing computing allocation for faculty at Illinois will be leveraged while seeking external fellowships and cost-sharing agreements with specific programs such as NIH’s Advanced Research Projects Agency for Health, NSF Small Business Innovation Research, Technology, Innovation and Partnerships, Formal Methods in the Fields; Bill & Melinda Gates Foundation; Intel; IBM; Amazon; and Mayo Clinic, as our PIs have been funded by and will seek additional funding from those organizations. We also plan to work with our clinical partners (pathai.com, franklin.ai) in Vietnam and the U.S. to leverage their pathologist network and labeled data for third-party evaluation. We will work with the U.S. Agency for International Development to support Vietnam veterans and affected Vietnamese in studying skin cancers and helping the treatment of Agent Orange’s effect. Finally, a dedicated team at the proposal development office at CSL/NCSA will support VinUni’s graduate students and PIs in this endeavor.
Digital Pathology Applications: Squamous cells on skin cancer (Mayo leads)
Cutaneous squamous cell carcinoma (cSCC) is the second most common skin cancer in the U.S., affecting over one million individuals annually and kills as many people per year as melanoma. In particular, Vietnam veterans who have been exposed to Agent Orange are particularly vulnerable [1]. Clinicians evaluate the risk of unfavorable outcomes such as metastasis and recurrence by examining tumor attributes like cellular differentiation and invasion depth by examining tissue images. Current staging systems, the American Joint Committee on Cancer (AJCC)-8, use clinical and histopathological risk factors to assess the risk of recurrence or metastasis. Our prior work has demonstrated that inadequate clinical or histopathological information results in understaging of 46% of high-risk.
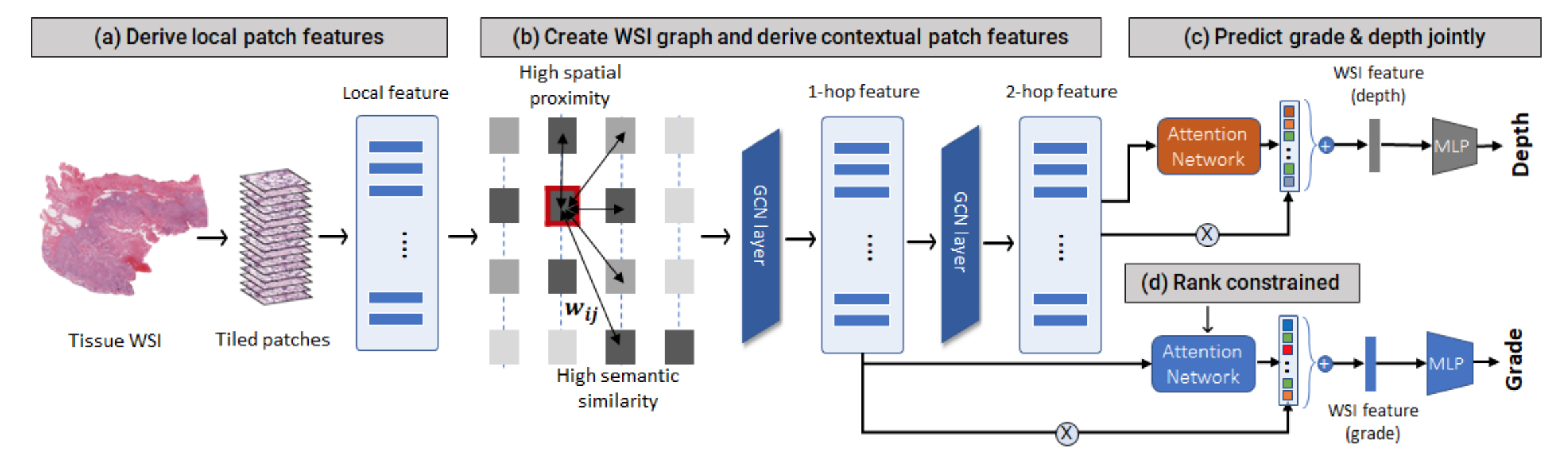
Figure 2: ML workflow analyzing tissue patches in collaboration with Mayo that NCSA’s supercomputer will accelerate.
cSCC tumors before surgery. For these reasons, current clinical/ histopathological staging systems for intermediate to high-risk cSCC are inadequate, and the true number of at-risk tumors without clear management guidance may be as high as 250,000. Moreover, the current practice for grading cSCC tumors through manual examination of whole slide images is inherently subjective and prone to inter-observer variability.
Preliminary results. Our research team has developed the first comprehensive AI-tool for cutaneous squamous cell carcinoma (cSCC) grading (differentiation prediction) and staging (depth prediction) using tissue images to assist pathologists in patient risk stratification (Fig. 2). The proposed approach uses weakly-supervised learning and does not require tedious pixel-level annotations in images. Using the multiple instance learning paradigm, the system predicts tissue tumor grade via four main steps. First, we divide the tissue into patches and extract local patch features using a self-supervised pre-trained encoder. Second, we define a graph on the tissue patches that captures both local (spatial) and non-local (semantic) dependencies between the patches. We derive multiscale features using self-attention-based graph convolution to incorporate contextual information. Third, we utilize attention-based patch feature aggregation to derive a WSI-level feature for grade classification. We augment the attention computation with a rank ordering mechanism that assigns higher weights to higher-grade tumor regions. Finally, we incorporate tumor depth as an auxiliary prediction task for regularizing attention to relevant tumor patches. Our initial results are promising, with the multimodal model achieving 79% accuracy on metastasis prediction with a 2% improvement by using both image and genomic data.
Proposed research. We will: 1) examine the complementary information between patch level H&E (structure and cell morphology) and corresponding patch level features associated with genetic changes to integrate multimodal imaging, genomic, and clinical features, and develop a multivariate Cox proportional hazards model for risk prediction. By leveraging clinical domain knowledge and accelerated computation on NCSA’s supercomputer, our model will integrate three key innovations to enable robust learning with limited tissue annotations and small imbalanced training data.
Rank-ordering: To emulate the ordinal grading protocol implicitly followed by pathologists, we introduce a two-part rank-ordering loss to train the attention network. It consists of (i) an interclass constraint, which compares patches from different grades and imposes greater attention on more severe tumor patches, and (ii) an intraclass constraint, which imposes greater attention on more likely patches within the same grade.
Tissue graph: Second, we demonstrate the effectiveness of combining local and non-local dependencies between tissue regions for grading. We define a graph network on the tissue image with patches as nodes and edges defined using a combination of spatial proximity and semantic similarity, i.e., patch feature similarity. Semantic similarity enables long-range relationship modeling extending beyond the immediate neighbors in WSI during graph convolution while incorporating spatial and semantic context to improve the localization and classification of higher-risk tumors (moderately and poorly differentiated).
Tumor depth as auxiliary task: Finally, we introduce tumor depth as an auxiliary training signal to enhance grade classification. Prior studies have shown that tumor grade is significantly associated with tumor depth. Well-differentiated cSCC tumors have lower depth, while poorly differentiated tumors invade deeper into the tissue. To capture the relationship between depth and grade, we develop a multitask framework that predicts depth and grade jointly, sharing the patch features between them. Our approach successfully localizes relevant tumor regions with 30-50% higher sensitivity than existing methods, and qualitative analysis reveals that the tumor region(s) localized by the model aligns well with pathologists’ tumor annotations.
In sum, to refine our multimodal approach, we will leverage Bayesian Factor Graphs defined based on biological interactions (gene pathways, tissue image dependencies) to derive modality-specific features capturing within-modality correlations. Our method is broadly applicable to other cancers as described below.
2. Osteosarcoma bone cancer (VinUni-Vinmec lead, NCSA/CSL support)
Osteosarcoma is a rare malignant bone tumor with a predilection for adolescents and young adults, driving $12.6 billion oncology cost [13]. In developing countries such as Vietnam with limited care, bone cancer significantly affects the activities of daily living (ADL) and quality of life (QOL) in cancer patients due to skeletal-related events. We have worked with Dr. Tran Trung Dung, who performed innovative world-first surgeries in Vietnam, such as replacing the entire arm bone [14, 15], to gain clinical insights into a longitudinal, nationwide dataset of more than 100 osteosarcoma cancer patients in Vietnam.
Preliminary results. We performed initial data collection, involving the compilation of medical records and multimodal images, including X-rays and MRIs, from more than 100 bone cancer patients. Fig. 3 summarizes key distributions of the dataset from Vinmec, particularly tumor anatomical location, diagnosis time, treatment plan including surgical distribution, and patient’s survival time. Results show major challenges: 1) accessible, low-cost, preventative screening diagnostic equipment such as X-Rays are prohibitively expensive for a large population; 2) characterization of country-wide patients to advance our understanding of tumor progression is limited; and 3) advanced (AI) tools to support doctors in pre-surgery planning, patient outcome estimations, and post-surgery recovery management are lacking. To analyze the survival rates of patients, we used Cox proportional hazards regression. This semi-parametric model assesses the impact of covariates on the hazard rate while assuming that the hazard ratio is constant over time [16].
Proposed research. We will: 1) systematically characterize the screening, pre-surgery, rare case studies, and recovery effectiveness of bone cancer patients in Vietnam; 2) use counter-factual reasoning and domain knowledge to support symptoms diagnosis, reduce uncertainty in patient outcome, and predict survival rate for each treatment plan. Specifically, we will employ Bayesian causal analysis,
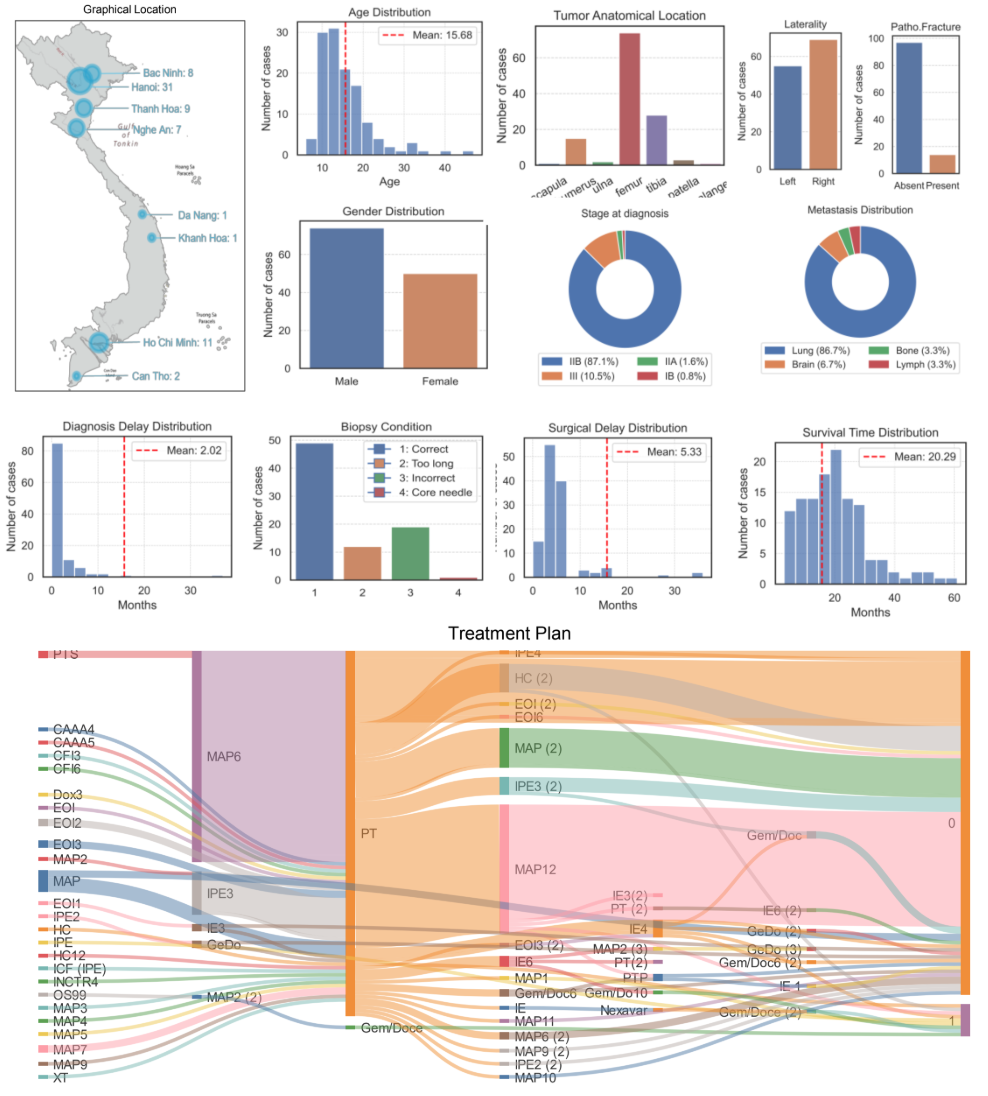
Figure 3: Vietnamese Data on Osteosarcoma Cancer (VinUni student Chi Phan collaborated with Dr. Tran Trung Dung at VinMec)
such as counterfactual reasoning [17], to unravel the complex links between key risk factors, laboratory test results, treatment plans, and patient survival outcomes. By integrating data from other applications on a supercomputing platform that our PI has access to, our research aims to identify prognostic factors that are indicative of tailored treatments for individual patients, ultimately enhancing survival rates and improving the post-treatment quality of life. We will borrow the features learned by the previous squamous cell model to enhance diagnostic decision-making, providing valuable morphological insights that can be integrated with disease-specific gene-expression panels (developed by our research team) for outcome prediction.
3. Polyp Surveillance: from AI-Multiomics Digital Pathology (CSL/UI Health lead) & to Soft Robotics Surgery (VinUni-Vinmec lead)
Colorectal cancer (CRC), the second leading cause of cancer-related deaths in the U.S., disproportionately impacts racial and ethnic minorities, including Vietnamese Americans. Colorectal cancer develops via abnormal growth of the colon epithelium, leading to adenomatous polyp. Up to 60% of all individuals undergoing colonoscopy will have at least one polyp, and 48% of advanced polyps recur at the same location within 3 years of the first polyp removal. While most precancerous adenomas (95%) do not progress to cancer, they have the potential to transform into cancer, given enough time to grow. The current state-of-the-art colonoscopy continues to be the foremost method for screening colorectal cancer (CRC) and is recognized as the gold standard in population-based screening programs worldwide. It’s important to note that nearly 60% of CRC-related deaths could be averted through effective screening. However, colonoscopy attendance rates can be impacted by concerns like discomfort, fear of pain, and feelings of embarrassment or anxiety regarding the procedure. In addition to these challenges, the emergence of new communicable diseases on a global scale poses potential threats to the seamless functioning of contemporary gastrointestinal endoscopy centers. Despite rigorous device sterilization procedures, the risk of infections related to endoscopic devices remains a significant concern, with the potential to cause substantial harm to patient’s health and well-being.

Figure 4: Polyp Surveillance: from AI-Multiomics Digital Pathology to Soft Robotics Surgery
Preliminary results. We: 1) identified specific molecular features that predict the transformation potential of a polyp that will assist the understanding of pathologists and 2) developed a prototype soft robotic system for in situ 3D bioprinting and endoscopic surgery [18] developed by Dr. Thai at VinUni (Fig. 4). The identified molecular attributes distinguish non-recurring polyps from recurring polyps curable by colonoscopy. Molecular changes have been shown to manifest in histologic features in CRC. Based on preliminary analysis and existing research linking histologic features with molecular changes, we hypothesize that histologic features with multi-omics will allow us to decipher disease mechanisms that determine polyp outcome and cancer and recurrence potential.
Proposed research. Multimodal AI will discern the underlying correlation between histology and molecular features and work with soft robotics to increase patients’ survival rates as follows.
- Evaluation of endoscopic and histologic/molecular features would reduce the disruption of a polyp removal process and transfer to specimen containers for pathologist review. The correlation of molecular events with distinct clinical behavior can be used to bridge knowledge gaps on polyp recurrence or cancer progression. Identifying molecular alterations will lead to mechanistic insights and candidates for polyp vaccines or chemopreventives. The technology can be applied to normal colons to predict the development of polyps in the future, thus providing individualized recommendations regarding cancer screenings and polyp surveillance, leveraging foundational models developed jointly with the skin cancer project.
- Real-time AI-enabled histologic and molecular intensified assessment, accelerated by supercomputers, will lead to a novel approach similar to MOHS (microscopically controlled surgery) in skin cancer management. At the time of initial polypectomy, the resection margins would be verified as cancer-free and polyp-free; a resection margin can be obtained. A virtual histologic assessment tool integrated into the colonoscope would simplify polypectomy based on real-time polyp-free measurements. The technology would benefit early-stage colon and rectal cancer diagnosis, reducing patient risks, recovery times, and costs. We will realize microscopically controlled surgery via a soft robotics system as follows.
- Low-cost, disposable robotic endoscopic arm. The adoption of disposable devices stands as a compelling incentive for the future advancement of colonoscopy, offering an innovative solution to the existing challenges. Dr. Thai will extend his novel, low-cost (up to 100x cheaper than existing commercial solutions), disposable robotic endoscopic arm designed for both gastrointestinal diagnosis and the detection of gastrointestinal cancer. This 3-degree-of-freedom (3-DOF) endoscopic arm operates through the use of three soft fabric bellow actuators thoughtfully arranged in a parallel configuration at the vertices of an equilateral triangle, allowing for seamless omnidirectional movement. The endoscopic arm is seamlessly integrated into a long, flexible catheter, serving as the body of the endoscope. To further enhance the diagnostic capabilities, the developed AI-based computational pathology will leverage data obtained through this endoscopic arm for training robust models, subsequently integrating these capabilities into the endoscopic arm as a powerful diagnostic tool.
Summary: Mayo’s extraordinary expertise in gastroenterology and colon cancer and their valuable endoscopic and histological omics data, combined with VinUni and Illinois’ experience in developing AI/ML will assist pathologists’ understanding of cancer progression, boost their productivity, determine when a polyp is at risk for recurring or transforming to cancer, prioritize patients who need a timely colonoscopy, and actuate precision surgery via soft robotics methods. Healthcare costs are realized by reducing the steps to reaching a diagnosis, and our research will enrich scientific knowledge through open data& models in the key milestones (Fig. 5).
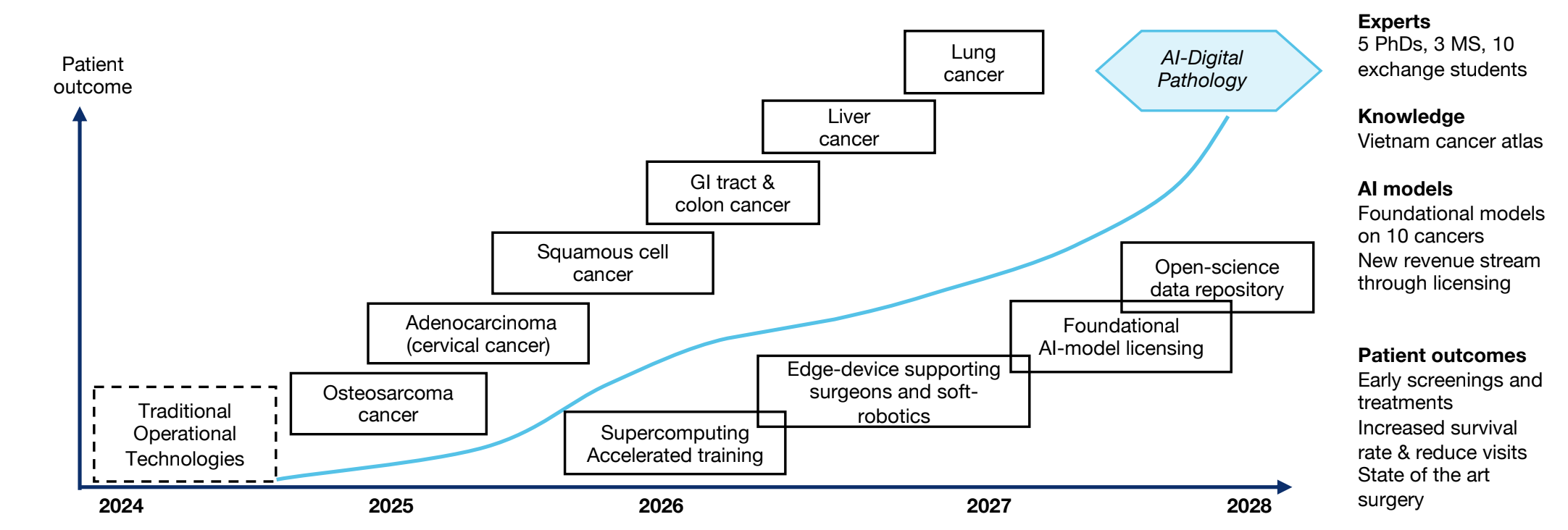
Figure 5: Milestones. Project will start the second half of August 2024 (24H2) – August 2028 (28H2)
References
- Innumerable squamous cell carcinomas in vietnam war veteran exposed to chemical defoliants – pmc. https://www.ncbi. nlm.nih.gov/pmc/articles/PMC6715125/#:˜:text=Agent%20Orange%20exposure%20has%20been,veterans% 20of%20the%20Vietnam%20War.&text=Further%20research%20is%20needed%20to,primary%20therapy%20for% 20localized%20SCCs. (Accessed on 10/29/2023).
- Nsf awards ncsa $10 million for deployment of delta – ncsa. https://www.ncsa.illinois.edu/ nsf-awards-ncsa-10-million-for-deployment-of-delta/. (Accessed on 10/31/2023).
- Home – ncsa. https://www.ncsa.illinois.edu/. (Accessed on 10/31/2023).
- Brett Bode, Michelle Butler, Thom Dunning, Torsten Hoefler, William Kramer, William Gropp, and Wen-mei Hwu. The blue waters super-system for super-science. In Contemporary high performance computing, pages 339–366. Chapman and Hall/CRC, 2017.
- Subho S Banerjee, Arjun P Athreya, Zbigniew Kalbarczyk, Steven Lumetta, and Ravishankar K Iyer. A ml-based runtime system for executing dataflow graphs on heterogeneous processors. In Proceedings of the ACM Symposium on Cloud Computing, pages 533–534, 2018.
- The cancer genome atlas program (tcga) – nci. https://www.cancer.gov/ccg/research/genome-sequencing/tcga. (Ac-cessed on 10/31/2023).
- Sean Lie. Cerebras architecture deep dive: First look inside the hw/sw co-design for deep learning: Cerebras systems. In 2022 IEEE Hot Chips 34 Symposium (HCS), pages 1–34. IEEE Computer Society, 2022.
- Yogatheesan Varatharajah, Min Jin Chong, Krishnakant Saboo, Brent Berry, Benjamin Brinkmann, Gregory Worrell, and Ravishankar Iyer. Eeg-graph: a factor-graph-based model for capturing spatial, temporal, and observational relationships in electroencephalograms. Advances in neural information processing systems, 30, 2017.
- Yurui Cao, Phuong Cao, Haotian Chen, Karl M Kochendorfer, Andrew B Trotter, William L Galanter, Paul M Arnold, and Ravishankar K Iyer. Predicting icu admissions for hospitalized covid-19 patients with a factor graph-based model. In Multimodal AI in healthcare: A paradigm shift in health intelligence, pages 245–256. Springer, 2022.
- Subho S. Banerjee, Saurabh Jha, Zbigniew Kalbarczyk, and Ravishankar K. Iyer. Bayesperf: Minimizing performance monitoring errors using bayesian statistics. In Proceedings of the 26th ACM International Conference on Architectural Support for Programming Languages and Operating Systems, ASPLOS ’21, page 832–844, New York, NY, USA, 2021. Association for Computing Machinery.
- Subho S. Banerjee, Saurabh Jha, Zbigniew T. Kalbarczyk, and Ravishankar K. Iyer. Inductive-bias-driven reinforcement learning for efficient schedules in heterogeneous clusters. In Proceedings of the 37th International Conference on Machine Learning, ICML’20. JMLR.org, 2020.
- Mary Pietrowicz, Amrit Kamboj, Manoj Yarlagadda, Kevin Buller, Keiko Ishikawa, Diana Orbelo, and Cadman Leggett. Speech-based biomarkers for automatic detection of gerd: A pilot study. Acoustical Society of America Journal, 151(4):A63–A63, 2022.
- Kathy L. Schulman and Joseph Kohles. Economic burden of metastatic bone disease in the u.s. Cancer, 109(11):2334–2342, 2007.
- Dung Tran Trung, Sang Nguyen Tran Quang, Hieu Pham Trung, Nam Vu Tu, Nang Vo Sy Quyen, Thanh Tran Duc, Nguyen Tien Dung, Tran Thiet Son, Pham Thi Viet Dung, and Nguyen Van Truong. Partial replacement of pelvis with the hip joint in osteosarcoma treatment: A case report. Annals of Medicine and Surgery, 70:102812, 2021.
- World first as vietnamese patient has entire arm bone replaced. https://vietnamnews.vn/society/1073265/ world-first-as-vietnamese-patient-has-entire-arm-bone-replaced.html. (Accessed on 10/31/2023).
- David R Cox. Regression models and life-tables. Journal of the Royal Statistical Society: Series B (Methodological), 34(2):187–202, 1972.
- Saurabh Jha, Shengkun Cui, Zbigniew Kalbarczyk, and Ravishankar K Iyer. Watch out for the safety-threatening actors: Proactively mitigating safety hazards. arXiv preprint arXiv:2206.00886, 2022.
- Mai Thanh Thai, Phuoc Thien Phan, Hien Anh Tran, Chi Cong Nguyen, Trung Thien Hoang, James Davies, Jelena Rnjak-Kovacina, Hoang-Phuong Phan, Nigel Hamilton Lovell, and Thanh Nho Do. Advanced soft robotic system for in situ 3d bioprinting and endoscopic surgery. Advanced Science, 10(12):2205656, 2023.
