Wastewater Epidemiological Surveillance in Vietnam
Motivation
Since the beginning of civilization, infectious disease pandemics have periodically threatened humanity. Newly emerging and re-emerging pathogens and antibiotic resistance continue to challenge global public health in the 21st century. Vietnam has successfully contained viral pandemics of SARS in 2002 (CDC), influenza H1N1 in 2009 (Let et al., 2014), and the first wave of COVID-19 in 2020 (Cobelens & Harris, 2021; Pham et al., 2021). Despite the availability of vaccines since January 2021 in the US and Europe and gradually elsewhere, emerging new variants of SARS-CoV2, which may have better infectivity, transmissibility, and immune evasion, again threaten global public health (Harvey et al., 2021; Lazarevic et al., 2021). Indeed, the rise of the highly contagious Delta variant has proved much harder to contain worldwide, including in Vietnam (Hoang et al., 2021). Monitoring the advent and prevalence of variants of concern (VOCs) as well as variants of interest (VOIs) in a community can help local health authorities to make informed decisions regarding public health (Grubaugh et al., 2021). Since the onset of COVID-19, pre-sewage treatment wastewater sampling has been harnessed as a non-evasive large-scale pathogen surveillance tool (Peccia et al., 2020; and Saththasivam et al., 2021). A dashboard now shows the applicability of global COVID-19 surveillance using wastewater (COVIDPoops19 (arcgis.com)). Wastewater surveillance complements clinical diagnosis because early detection of transmission by asymptomatic infected individuals is achieved without excessive resources and labour required for frequent human testing. The establishment of infrastructure for wastewater-based SARS-CoV2 surveillance will serve as an entry point for monitoring other infectious agents, antimicrobial resistance (AMR) genes, and even chemicals or biomarkers that humans excrete during the course of infectious or metabolic diseases.
I. Research Questions
While a pandemic is global, its transmission is local. Therefore, surveillance should be designed around local contexts. Besides COVID-19, antimicrobial resistance (AMR) is a worsening silent pandemic, as predicted by the WHO. Increased usage of prescribed antibiotics for COVID-19 patients has been reported (Mahoney et al., 2021; Patel et al., 2021). COVID-19 is likely to exacerbate AMR worldwide, especially in Vietnam where direct accessibility of consumers to pharmacies has led to the overuse and abuse of antibiotics (Mao et al., 2015).
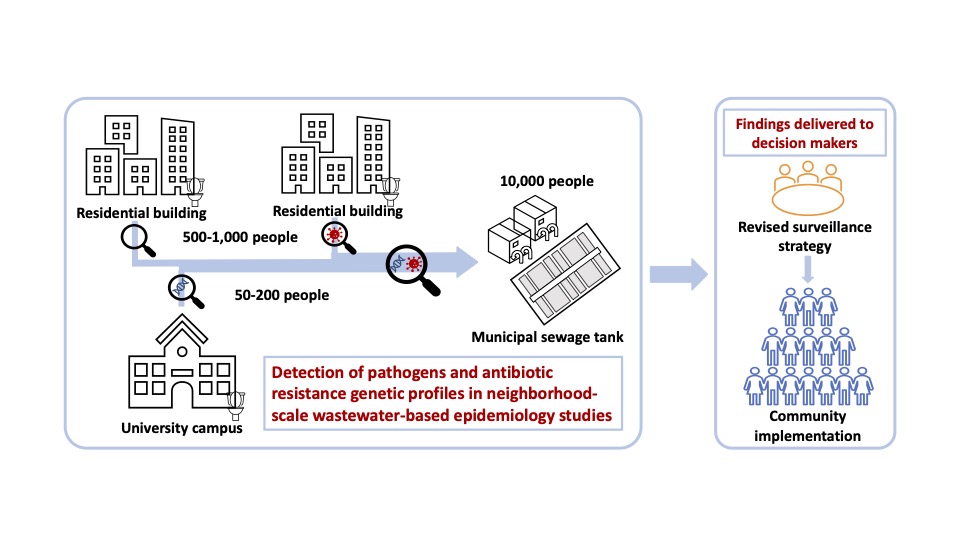
How to identify sentinel sites for wastewater surveillance is the most important research question. The identification of sentinel sites and the frequency of sampling depend on specific, actionable surveillance goals. These require a different approach in a public health crisis to endemic conditions. Examples of key research questions for consideration are below. The answers to these will require the integration of researchers from various disciplines in the Smart Health Center.
1) What factors (past infectious data, seasonality, human mobility, human behavior, population density, number of houses served by a sewage pipe, and others) guide sentinel site selection?
2) How do the natural history of pathogens, their life cycles, and disease transmission influence the locations and frequency of sampling?
3) How to establish a surveillance system that can capture an outbreak’s evolution and thereby help prevent its escalation to an epidemic, pandemic and eventual endemicity, and evaluate the effectiveness of public health interventions?
4) How to identify hotspots of infection and transmission routes (large apartment complexes, airports, public transportation hubs, schools, places of public gatherings, etc.)?
5) Can we use artificial Intelligence-assisted tools to assess potential transmission and to identify sentinel sites?
II. Approach
1) Overview:
Finding answers to the above questions requires a functional surveillance system informed by modeling of transmission. Therefore, we propose immediately setting up and testing a sewage surveillance system for each high-rise residential building in the Times City urban complex located near Vinmec, Hanoi, and developing a hierarchy of epidemiological models to investigate the spread of infection in this community. These models and the sewage surveillance system will be complemented by human testing. Once these initial trials are completed, we will expand the surveillance systems and develop long-term research projects for PhD students and postdoctoral fellows, with supervisory teams of faculty from each university as appropriate, to answer the aforementioned questions.
The initial stage consists of two aims to be completed within one year. In aim 1, we propose to establish a sewage surveillance system capable of quantifying SARS-CoV2 and its variants of concern. We emphasize that quantification of virus variants is crucial for the following reason. Variants have different transmission capabilities. A variant with high transmission capability, such as Delta, provides an early warning sign for potential widespread transmission in the community. Quantitative results (instead of positive/negative) are also important as the trend in genome concentration enables prediction of future transmission levels. In aim 2, we propose to develop infection models that account for: (1) Heterogeneous biological and social susceptibility to the disease; (2) Emergence of new SARS-CoV2 variants of concern that might exhibit enhanced transmission; and (3) vaccination coverage. This adaptive modeling strategy will be calibrated continuously using data available from the sewage surveillance system and additional statistics on hospitalization, ICU occupancy, and cause of death. This approach will enable a real-time evaluation of epidemic status in the community, short-term prediction of the course of the epidemic, and guidelines for longer-term mitigation. Aim 3 will focus on understanding the prevalence and emergence of AMR in these communities. Aim 3 will be a research topic for several PhD students.
Aim 1: Sewage surveillance system
a) Sampler deployment:
Sewage from each building will be collected by an automated sampler connected to a sewer pipe (Gibas et al., 2021). We propose that wastewater sampling at these sentinel sites should be composite instead of a one-time grab sampling. Composite sampling means taking a small volume of sewage from each pipe (e.g. 10-200 mL) every hour for 24 to 72 hrs. This method allows the collection of samples over time to capture the unpredictable release of human body waste. For SARS-CoV2, sampling on alternate days is necessary to identify positive individuals with more than 90% sensitivity during their infectious period in individual-level PCR testing (Smith et al., 2021). It, therefore, should provide adequate sensitivity to detect cases using wastewater. On the other hand, sample collection every week for 3 days each has also been implemented successfully by Helen Nguyen’s team to provide 7 to 14 days’ early warning sign in Champaign- Urbana. Sampling will be conducted for at least one year and will continue depending on the findings and the research questions initiated based on the findings.
b) Sample processing and analysis
Upon the arrival of composite sewage samples at the Vinmec laboratory, we will gently remove the supernatant and add 200 μL of bovine coronavirus (BCoV) to the remaining solution (about 50 mL) to serve as an internal virus control. After centrifugation, the liquid is removed, and the solid part is subjected to viral RNA extraction. The extracted RNA will be used to quantify the genome concentration of SARS-CoV2 in each sewage sample. We will apply a multiplex quantitative RT- qPCR assay developed by Helen Nguyen’s group to quantify the concentration of N1 gene (a conserved gene of SARS-CoV2) and target specific mutation in the viral genome for VOCs. Our analysis of viral sequences from GISAID reveals that the Delta variant occurred in Vietnam in April 2021 and quickly replaced the Alpha variant as the predominant type. The same samples will also be analyzed for antibiotic resistance genes (ARGs) described in Aim 3.
Aim 2: Epidemiological model development for COVID-19 transmission
For this proposal, we plan to model the Time City community as a collection of different spatial regions. Each region will be described by an instance of our recently developed behavior-informed epidemiological models. Interactions between different regions will be modeled using an interaction matrix that accounts for human mobility data and socio-economic profile. Of particular relevance, wastewater surveillance and associated targeted testing will be modeled in this framework as part of the adaptive mitigation. Virus levels detected in wastewater in a given region will be mapped to an initial number of infections that will drive the model. Targeted testing will identify infected but likely still asymptomatic individuals, enabling their isolation, thereby breaking the transmission chain earlier. Changes in human behavior in response to rising or declining case numbers, including working from home, self-isolation, or variation in commuting trends, affect different population groups’ susceptibility. Additional modulation of susceptibility arises from varying rates of COVID-19 vaccine uptake. Finally, stochastic social gathering activities associated with parties, family visits, festivals, and other socialization modes will contribute to the emergence of a variable infection rate over different time scales. Data from social media, surveys, newspapers, smartphone, and traffic reports will inform our quantitative modeling of this human behavior component.
Model predictions will inform testing deployment, increase testing frequency to target clusters of infection, and spatial distribution of wastewater sampling to preemptive action on propagating infection waves. The multi-community spatial-temporal nature of our model balances accuracy with computational cost and provides sufficient flexibility to respond to real-time data, adapting accordingly to provide an ensemble forecast that guides decision-making. Ahmed Elbanna has expertise in epidemiological modeling at different population and time scales, including the state of Illinois (Wong et al., 2020), a large public university with 30,000 student population, heterogeneous social behavior, and different non-pharmaceutical mitigation strategies (Ranoa et al., 2021; Tkachenko et al., 2021).
Aim 3: AMR surveillance and correlation with antibiotic usage
This aim will connect the prevalence and emergence of ARG with antibiotic usage in Times City and other studied communities. We will encourage residents to use an app developed by Minh Do’s team that reminds these residents to take their medicine, including antibiotics. We will receive aggregated data from the app on the usage of antibiotics in the same communities where the sewage was collected and analyzed. We will then detect ARGs within the entire microbial community and analyze the whole-genome sequence of enteric bacteria isolated from sewage samples, such as E. coli or the Enterobacteriaceae family, to investigate horizontal gene transfer. We will first screen 320 genetic markers of ARGs and mobile gene elements (MGEs) for the entire bacterial DNA extracted from the sewage samples using the methods developed previously by Helen Nguyen’s group (Mao et al., 2021a & b). This screening will provide information on the overall presence of ARGs. However, it does not give information on horizontal gene transfer (HGT), which is needed to predict whether ARGs can be transferred to a powerful pathogen. We propose to study HGT using two methods. The first method is a CRISPR- based method called FLASH to design guide RNA based on ARG screening results and ARG databases such as CARD and ResFinder, then amplify the target ARG sequences accordingly for sequencing (Quan et al., 2019). The second method involves whole-genome sequencing of bacterial isolates using Illumina sequencing followed by bioinformatics tools such as Spine, gplas and ClustAGE (Ozer et al., 2014; Ozer, 2018; Arredondo-Alonso et al., 2020). The genome of selected bacteria isolates will be sequenced by long-range sequencing to further investigate the relationship of the CRISPR arrays and the plasmids among those isolates (Lam & Ye, 2019). The sequencing results will be analyzed using various computational tools to determine HGT. The combination of aggregated data from Minh Do’s team and the study of HGT will give information about the selective pressure on the emergence of AMR in response to human practices of taking medicines to treat infectious diseases, particularly antibiotics for bacterial infection, but including public health threats that are viral, fungal or parasite in origin.
III. Intellectual Merits
In the short-term, the project will determine the prevalence of SARS-CoV2 in the selected communities. In the long-term, the project findings will give valuable insights into understanding the evolution of AMR in response to human behavior and community practices (taking unprescribed drugs and not completing the course) and medical practices (overprescription of antibiotics). The project will also develop a non-invasive, privacy- preserved tool for epidemiological surveillance for pathogen transmission in local communities. While a pandemic is global, its transmission is local and should be studied in a local context.
1. Team
- Helen Nguyen, Ivan Racheff Endowed Professor of Civil and Environmental Engineering, PhD, Thanh Huong Nguyen | Civil & Environmental Engineering | UIUC (illinois.edu),
- Ahmed Elbanna, Donald Biggar Willett Faculty Fellow, Associate Professor of Civil and Environmental Engineering, PhD, Ahmed Elbanna | Civil & Environmental Engineering | UIUC (illinois.edu)
- Minh Do, Thomas and Margaret Huang Endowed Professor of Electrical and Computer Engineering, PhD, Minh N. Do (illinois.edu), Honorary Vice Provost of VinUni.
- Andrew Taylor-Robinson, Professor of Microbiology & Immunology, Andrew W. Taylor-Robinson, PhD – VinUni.
- Maurizio Trevisan, Professor and Dean, College of Health Sciences, Maurizio Trevisan, MD – VinUni.
- Nguyen Xuan Hung, Director, Center of Applied Sciences, Regenerative Medicine and Advance Technologies, Vinmec & Affiliate Faculty, College of Health Sciences, Nguyen Xuan Hung, PhD – VinUni,
- Le Cu Linh, Assoc. Prof and Vice Dean, College of Health Sciences, Le Cu Linh, MD, PhD – VinUni.
- Huynh Dinh Chien, Professor, College of Health Sciences, Huynh Dinh Chien, MD, PhD – VinUni
2. Project Plan:
Year 1: Aim 1 – surveillance system set-up, site investigation, and lab set-up will start in Jan. Sampling will start in June for one year by undergraduate, MS, and PhD students of VinUni co-advised by our team. Aim 2 – Students will be trained and start working also in Year 1. Aim 3 will start in Years 2-5. Samples will be collected for the first year, unless there is a need to extend surveillance for SARS-CoV2. A postdoc or student from Helen Nguyen’s lab will come in June of Year 1 to help train Vin Uni students.


